Di Meng
Computer Science PhD
University College Dublin
di.meng@ucdconnect.ie
Biography
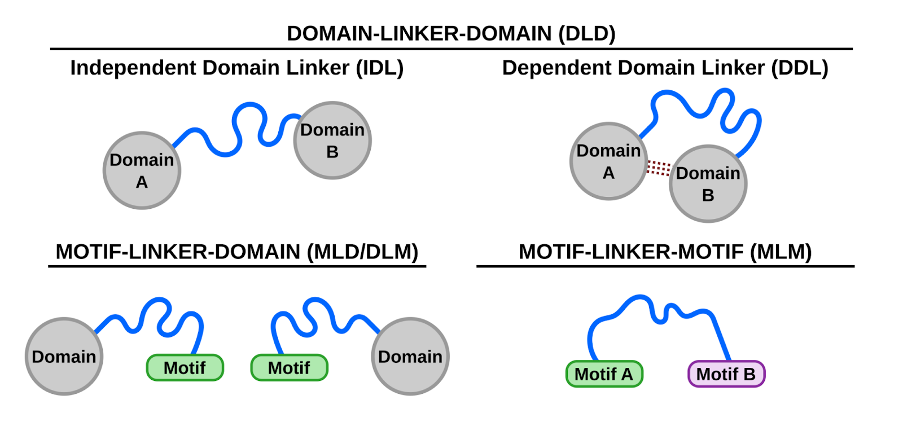
I am a PhD researcher in computational biology, specializing in intrinsically disordered protein (IDP) structure and function prediction. Since 2021, I have been pursuing my PhD at the School of Computer Science, University College Dublin, Ireland, under the supervision of Dr. Gianluca Pollastri. My research focuses on applying deep learning and bioinformatics to enhance the analysis and prediction of protein disorder. I aim to complete my PhD by September 2025.
Before my PhD, I worked in various software development and data science roles across multiple companies, gaining experience in algorithm development, machine learning, and computational modeling.
Beyond research, I have a strong interest in cooking and enjoy running, casual exercise, and Yoga.
Interests
- Deep Learning
- Bioinformatics
- Debugging
- Yoga
- Running
Education
-
PhD in Computer Science, 2021-2025
University College Dublin, Ireland
-
Visiting Scholar, 2024
University of Padova, Italy
-
Visiting Scholar, 2022
National University of General San Martín, Argentina
-
Master in Computer Science, 2020
University College Dublin, Ireland